RAGs differ from standard LLMs because of the knowledge base that enriches user queries. This knowledge base acts as a repository of information that can be accessed and searched when a user queries the LLM on a specific topic. The higher the quality of this knowledge base, the better the LLM's final response will be. This is because the LLM will have access to more relevant and accurate information during the answer-generation process. Therefore, this article aims to build a solid and comprehensive knowledge base for our RAG system.
How to Create a Vector Database
Data preprocessing was the focus of the previous article in this series although we provided a brief overview of vector databases. For context, let's redefine vector databases for those already familiar with data preprocessing who may have skipped the previous article.
A vector database is a specialized type of database designed to store and manage data in vector format. These databases are optimized for efficiently handling high-dimensional data and are built to scale with large datasets, often containing millions or even billions of vectors. They are also performance-oriented, frequently using hardware acceleration like GPUs to facilitate rapid data retrieval. The main function of vector databases is to conduct similarity searches. In this process, the system quickly finds vectors that are most similar to a given query vector. This is achieved by calculating metrics such as Euclidean distance or cosine similarity.
Vector database choices are abundant. Chroma database was selected for a few specific reasons, the most significant being its integration with Langchain. Since we plan to develop our RAG system using both Langchain and LlamaIndex, it is crucial to choose a vector database that has already been proven to work seamlessly with these tools. By selecting such a database, we eliminate concerns about compatibility. This allows us to focus on making sure the system is intuitive and user-friendly, instead of worrying about integration issues.
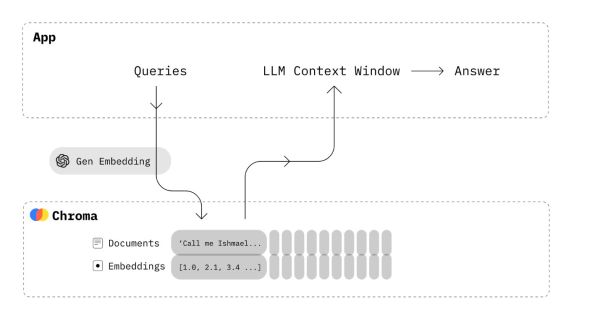
What Is Chroma Database
Chroma is an open-source vector database, that emphasizes efficiency and simplicity. It gives users the tools they need to:
- embed documents and queries
- store embeddings together with their metadata
- search for embeddings similar to an input query embedding
Chroma integrates seamlessly into an RAG system. First, we select an embedding model, which is used to transform text data into vectors. Afterward, we convert all the documents in our knowledge base into vectors and store them in the Chroma database. When a user submits a query, we use the same embedding model to turn the query into a vector. This vector representation allows us to search for similar vectors within the Chroma database. Once similar vectors are identified, we extract the associated data from the database. This data is then combined with the original query to create an enriched query. The enriched query is used to obtain a response from a large language model (LLM). Chroma is specifically optimized for handling these embeddings. It offers a user-friendly API that facilitates efficient searches, even with large databases.
- How to Create Custom Word Embeddings Using Gensim
- Word Embeddings in Natural Language Processing: The Complete Guide
Article continues below
Want to learn more? Check out some of our courses:
What Do We Need From Our Database
When creating a database, it is straightforward to convert all the data stored in a specific location on our computer into vectors. These vectors can then be stored in a database. However, this simplistic approach overlooks several key factors necessary for building an efficient RAG system.
First, after constructing the database from data stored on our computer, typically in a directory, we must ensure there are no duplicates. This requires implementing a system to track whether a file already exists in the database. Specifically, since we convert chunks created during preprocessing into vectors, it is essential to ensure that all chunks in the database are unique.
Second, we need the ability to update our database. This means we should be able to scan a directory and identify new files. We also need to add only those new files to the vector database, ignoring old files that are already stored.
Third, if a user replaces a file in the directory with a newer version, we need to handle this appropriately. Typically, this involves deleting the outdated information from the database and replacing it with the new data. This is a simple process if the user indicates a new version, such as by appending “v2” to the file name. However, most users will likely overwrite the original file with a new file of the same name. As a result, this requires a more nuanced approach to detecting and managing updates.
Let’s start building our database, and handling all of the aforementioned problems as we go. The entire process, start to finish, will consist of the following steps:
- preprocess data by loading in documents and splitting them up into chunks
- assign unique identifiers to chunks and create embeddings for them
- populate our database with those embeddings
How to Preprocess Data
The previous article of this series covered the first step: loading documents and dividing them into chunks. To accomplish this, we created two functions:
- load_documents()
- split_documents()
While the functions can remain as they are, it is a better idea to create a DocumentProcessor class. This class allows us to create a Python object that manages data loading and chunking. Let's implement that before we proceed with building our database.
from langchain.text_splitter import RecursiveCharacterTextSplitter
from langchain_community.document_loaders import PyPDFDirectoryLoader
class DocumentProcessor:
"""
A class to load and split documents using a recursive character splitter.
Attributes:
data_path (str): The path to the directory containing the documents.
splitter_type (str): The type of splitter to use ("recursive").
chunk_size (int): The size of each chunk (default is 800 for recursive).
chunk_overlap (int): The overlap between chunks (default is 80 for recursive).
Methods:
load_documents():
Loads documents from the specified directory.
split_documents(documents):
Splits the provided documents using the specified splitter type.
process_documents():
Loads and splits documents, returning the split document chunks.
"""
def __init__(self, data_path, splitter_type="recursive", chunk_size=800, chunk_overlap=80):
"""
Initializes the DocumentProcessor with the specified parameters.
The parameters are the directory path, splitter type, chunk size, and chunk overlap.
Args:
data_path (str): The path to the directory containing the documents.
splitter_type (str): The type of splitter to use ("recursive").
chunk_size (int): The size of each chunk (default is 800 for recursive).
chunk_overlap (int): The overlap between chunks (default is 80 for recursive).
"""
self.data_path = data_path
self.splitter_type = splitter_type
self.chunk_size = chunk_size
self.chunk_overlap = chunk_overlap
def load_documents(self):
"""
Loads documents from the specified directory and its subdirectories.
Returns:
generator: A generator yielding loaded documents.
"""
return PyPDFDirectoryLoader(self.data_path).lazy_load()
def split_documents(self, documents):
"""
Splits the provided documents into chunks using the specified splitter type.
Args:
documents (list[Document]): A list of Document objects to be split.
Yields:
Document: Chunks of the original documents after splitting.
Raises:
ValueError: If an unsupported splitter type is specified.
"""
if self.splitter_type == "recursive":
text_splitter = RecursiveCharacterTextSplitter(
chunk_size=self.chunk_size,
chunk_overlap=self.chunk_overlap,
length_function=len,
is_separator_regex=False,
)
else:
raise ValueError("Unsupported splitter type.")
for document in documents:
for chunk in text_splitter.split_documents([document]):
yield chunk
def process_documents(self):
"""
Unified method for loading documents, splitting them, and returning chunks of data.
Returns:
generator: A generator yielding split document chunks.
"""
for document in self.load_documents():
yield from self.split_documents([document])
Our DocumentProcessor class includes the two already defined functions, as well as a third function called process_documents(). This function essentially combines the functionalities of the two earlier functions. It allows the user to fully preprocess the data with a single call.
How to Assign Unique Identifiers to Chunks and Create Chunk Embeddings
After chunking our documents, we need to complete two steps before creating and populating the database with embeddings. First, we must assign a unique ID to each chunk. This will prevent duplicates in the database and ensure that, regardless of how many times the user updates it, the database will only contain the files from the specified source directory. This approach will also manage scenarios where a new version of a file replaces an older version. Second, we need to take an embedding model and use it to create embeddings for our unique chunks.
How to Create and Assign Unique Identifiers to Chunks
There are many ways to ensure that some chunk is unique. Our multi-faceted approach will ensure that each chunk is uniquely identifiable, even if it originates from the same source or contains similar content to other chunks. To do so, let’s create a new function called chunk_id().
import hashlib
import uuid
def chunk_id(chunk):
"""
Generate a unique identifier for a chunk based on its source, page, content, and a UUID.
This function checks if the current chunk is from the same page as the previous by comparing the current page ID
with the last page ID. If they match, the current chunk index is incremented,
indicating another chunk from the same page. If not, it resets to 0 for a new page.
A unique identifier for the chunk is created by combining this page ID, a hash of the chunk's content,
and a randomly generated UUID.
Args:
chunk (object): The chunk object containing metadata and content.
Returns:
tuple: Contains the newly created chunk ID and the current page ID.
"""
source = chunk.metadata.get("source")
page = chunk.metadata.get("page")
current_page_id = f"{source}:{page}"
content = chunk.page_content
content_hash = hashlib.sha256(content.encode('utf-8')).hexdigest()
unique_suffix = uuid.uuid4()
full_chunk_id = f"{current_page_id}:{content_hash}:{unique_suffix}"
return full_chunk_id, current_page_id
This function ensures that each ID is unique by combining several elements:
- metadata of the chunk
- hash of the chunk content
- randomly generated UUID
Most well-structured PDF files include metadata that allows us to pinpoint the exact file each paragraph comes from and the specific page within that file. This applies to the chunks we created in the previous step as well. For each chunk, we know precisely which document it belongs to and on which page of that document it is located. We can use this information as the foundation for generating our unique ID.
After that, we can hash the content of the chunk. Hashing the content of a chunk involves converting the text within the chunk into a fixed-size string of characters. This would typically be a sequence of letters and numbers, which is unique to that specific content, using a so-called cryptographic hash function. By hashing the content of each chunk, we can generate a unique identifier that is both compact and difficult to reverse-engineer into the original text. This hashed value changes even if the chunk's content changes slightly, providing a reliable method for detecting modifications and ensuring data integrity.
The algorithm that I am using here is SHA-256 (Secure Hash Algorithm 256-bit). It is a cryptographic hash algorithm that detects very minor changes in the input data, and based on those changes generates a vastly different value. Because of that, it is computationally infeasible to find two different inputs that produce the same hash output. As a result, SHA-256 is highly reliable for ensuring data uniqueness and security.
In summary, by incorporating the hash of the content into the unique ID, we ensure that any slight difference in the content will produce a different hash. This means that even if a chunk comes from a document with the same name as another document and from the same page, it will still be uniquely identifiable. This allows us to easily distinguish between the two chunks.
Finally, the last thing we will do is randomly generate a unique UUID and add that as the last part to our unique IDs. UUID stands for Universally Unique Identifier. It is a 128-bit number used to uniquely identify information in computer systems. A UUID is typically represented as a string of hexadecimal digits, divided into five groups separated by hyphens.
UUIDs are designed to be unique across both space and time. In other words, they are unlikely to be duplicated even when generated independently on different systems or at different times. In the context of generating unique IDs for document chunks, we use a UUID to ensure that each chunk can be uniquely identified, even if other identifying information (such as the source, page number, or content hash) might not be enough to guarantee uniqueness.
With this, our unique identifiers should be bulletproof, and we should never run into a situation where we have the same ID for two different chunks. Now that we have prepared the chunk_id() function we can generate unique IDs for our chunks. To generate those IDs using the function and to assign them to our chunks we will create a new function and name it process_chunks(). This function will assign unique IDs to chunks using the previously defined chunk_id() function.
def process_chunks(chunks):
"""
Process and uniquely identify each chunk using metadata, content hash, a UUID,
and manage chunk indexing across pages.
This function iterates through a list of chunks, generating a unique identifier for each chunk using metadata,
content hash,and appending a UUID.
The identifier is stored in the chunk's metadata. It also tracks and updates the page continuity and
the index of chunks within their respective pages.
Args:
chunks (list): A list of chunk objects, where each chunk has metadata including "source" and "page",
and content accessible via `chunk.page_content`.
Yields:
object: The chunk with updated metadata containing a unique identifier.
"""
for chunk in chunks:
chunk_id_str, last_page_id = chunk_id(chunk)
chunk.metadata["id"] = chunk_id_str
chunk.metadata["content"] = chunk.page_content
yield chunk
How to Create Embeddings for Chunks
After preparing the chunks, we can now use an embedding model to generate an embedding for each chunk. An embedding is a vector that represents the data that the chunk contains. In this case, the information is stored in the chunk text and is what we will store in our vector database.
Creating embeddings is straightforward when using Langchain as the framework. It simplifies the process by providing built-in classes and functions that handle embedding generation. To create embeddings, you simply need to initialize and return an instance of the OllamaEmbeddings class. This class is designed to work seamlessly with Langchain. It manages the conversion of text data into dense vector representations that capture the semantic meaning of the content. It allows us to leverage the power of pre-trained embedding models which will handle the “heavy lifting” of converting text into embeddings.
Let’s create a simple function called get_embeddings(), that will leverage the power of a pre-trained model to get us embeddings that represent our chunks.
from langchain_community.embeddings.ollama import OllamaEmbeddings
def get_embeddings(embedding_model="nomic-embed-text"):
"""
Initialize and return an instance of the OllamaEmbeddings class.
This function creates an OllamaEmbeddings object using the "nomic-embed-text" model and returns it.
Returns:
OllamaEmbeddings: An instance of the OllamaEmbeddings class initialized with the "nomic-embed-text" model.
"""
return OllamaEmbeddings(model=embedding_model)
The default model our system will use is the nomic-embed-text pre-trained embedding model. This model is specifically designed for generating embeddings and often performs better at this task than some of the models provided by OpenAI.
How to Create a Database and Populate It with Embeddings
Finally, it is time to create a database and populate it with embeddings that represent our chunks. To do so, let's create a ChromaDatabase class. This is the code for creating that class:
from langchain_community.vectorstores import Chroma
class ChromaDatabase:
"""
A class to interact with the Chroma database for storing and updating document chunks.
Attributes:
database_path (str): The path to the Chroma database.
db (Chroma): An instance of the Chroma database with embeddings initialized.
Methods:
update_database():
Processes documents and updates the database with new chunks, removing duplicates and persisting changes.
"""
def __init__(self, database_path):
"""
Initialize the ChromaDatabase with a specified database path.
Args:
database_path (str): The file path to the Chroma database.
"""
self.database_path = database_path # Set the database path
# Initialize the Chroma database with the given path and embeddings function
self.db = Chroma(persist_directory=self.database_path, embedding_function=get_embeddings())
def update_database(self, data_path):
"""
Processes documents and updates the ChromaDatabase with the resulting chunks.
Removes old chunks if it runs into new version of a particular file.
Then, it verifies the content of the database by printing the number of documents
after the update.
Args:
data_path (str): The path to the folder containing the PDF files.
"""
# Process documents to get chunks
document_processor = DocumentProcessor(data_path=data_path)
chunks = document_processor.process_documents()
# Process chunks to generate unique IDs
chunks_with_ids = process_chunks(chunks)
# Retrieve existing items in the database and extract their IDs
existing_items = self.db.get(include=[]) # IDs are always included by default
existing_ids = set(':'.join(item.split(':')[0:3]) for item in existing_items["ids"])
print(f"Number of existing documents in database: {len(existing_ids)}")
new_chunks = []
prefixes_to_clear = set()
# Identify new chunks by checking if their IDs (without UUID) are not in the existing IDs
for chunk in chunks_with_ids:
id_without_uuid = ':'.join(chunk.metadata["id"].split(':')[0:3])
if id_without_uuid not in existing_ids:
new_chunks.append(chunk)
prefix = chunk.metadata["id"].split(':')[0]
prefixes_to_clear.add(prefix)
# Remove all existing chunks from the database that start with any of the prefixes in prefixes_to_clear
if prefixes_to_clear:
chunks_to_remove = [chunk_id for chunk_id in existing_items["ids"] if chunk_id.split(':')[0] in prefixes_to_clear]
if chunks_to_remove:
print(f"Removing {len(chunks_to_remove)} existing documents with matching prefixes")
self.db.delete(ids=chunks_to_remove)
# Add new chunks to database
if new_chunks:
print(f"Adding {len(new_chunks)} new documents")
new_chunk_ids = [chunk.metadata["id"] for chunk in new_chunks]
# Add new chunks to the database and persist the changes
self.db.add_documents(new_chunks, ids=new_chunk_ids)
self.db.persist()
else:
print("No new documents to add")
# Verify the database content
existing_items = self.db.get(include=[])
print(f"Documents in database after update: {len(existing_items['ids'])}")
The class is designed to handle interaction with a Chroma database, specifically for storing and updating document chunks with embeddings. The class implements two methods:
- __init__()
- update_database()
The __init__() method is the constructor method. This method takes advantage of the Chroma implementation available in Langchain. To create the database using that implementation, we first need to define the path where we want to create it. If the database has already been created, we specify its location. Additionally, we need to define the embedding function that will be used to create embeddings before storing them in the database.
The update_database() method is designed to update the contents of our database, utilizing all the functions and classes defined earlier. The process it follows is quite easy:
First, it processes documents located on the computer, specified via the data_path argument. This is done to create chunks of content using the DocumentProcessor class. After this, it assigns unique IDs to these chunks using the process_chunks() function.
Next, the method examines the Chroma database to retrieve the set of chunk IDs already stored there. If this is the first time update_database() is being run, this list will be empty. If it has been run previously, there will be a list of existing IDs. For now, the method focuses on the source and page of the chunks by parsing the IDs to exclude the UUID. This helps check for updates.
The method then compares the chunk IDs of new chunks with the existing ones, identifying which new chunks need to be added to the database. If any prefixes indicate older versions of files, the corresponding outdated chunks are removed from the database.
Finally, the new chunks are added to the database, and the changes are persisted.
Throughout this update process, various print statements provide feedback on the amount of chunks (documents) in the database before and after running the update_database() method. When you run the method for the first time, the initial count of documents will be zero. The next time you run it, this count will not be zero, as data will have already been stored in the database.
Here is an example of creating an instance of the database class, and updating its contents:
# Create database instance
db = ChromaDatabase("CHROMA")
# Update database
db.update_database("data")
In my setup, I have a directory named data located within the directory where I am running the code. This data directory contains the PDF files I want to chunk, embed, and store in my vector database. By executing the code above, I will first create an empty Chroma database. Afterward, I will populate it with embeddings derived from the files in the data directory.
After the initial run, the printed statements would likely be something like:
Number of existing documents in database: 0
No new documents to add
Documents in database after update: 118
This happens because, on the first run, the Chroma database is created but remains empty initially. The result is a jump from zero to 118. In my case, it is 118 because that is the number of chunks generated after processing my data. If I try to update the database immediately afterward, I get the following:
Number of existing documents in database: 118
No new documents to add
Documents in database after update: 118
As can be seen, the system effectively recognizes that it has already processed these chunks and will leave the database unchanged.
- Self-Attention in Natural Language Processing: The Complete Guide
- Natural Language Processing and its Applications in the Finance Sector
In this article, we revisited how vector databases work, discussed the Chroma vector database, and demonstrated how to create and update a Chroma vector database. With these steps, we are nearly finished setting up our RAG system. In the next article, we will implement the logic to integrate the user's original prompt with the context extracted from the vector database. This will conclude our series on RAG systems.